Project Overview
This project was my final-year dissertation, focused on classifying TDP-43 protein aggregation patterns in ALS using deep learning and explainable AI. The aim was to distinguish between three clinical categories: Control (Healthy individuals), Concordant (ALS with cognitive impairment), and Discordant (ALS without cognitive impairment), based on post-mortem immunohistochemistry images.
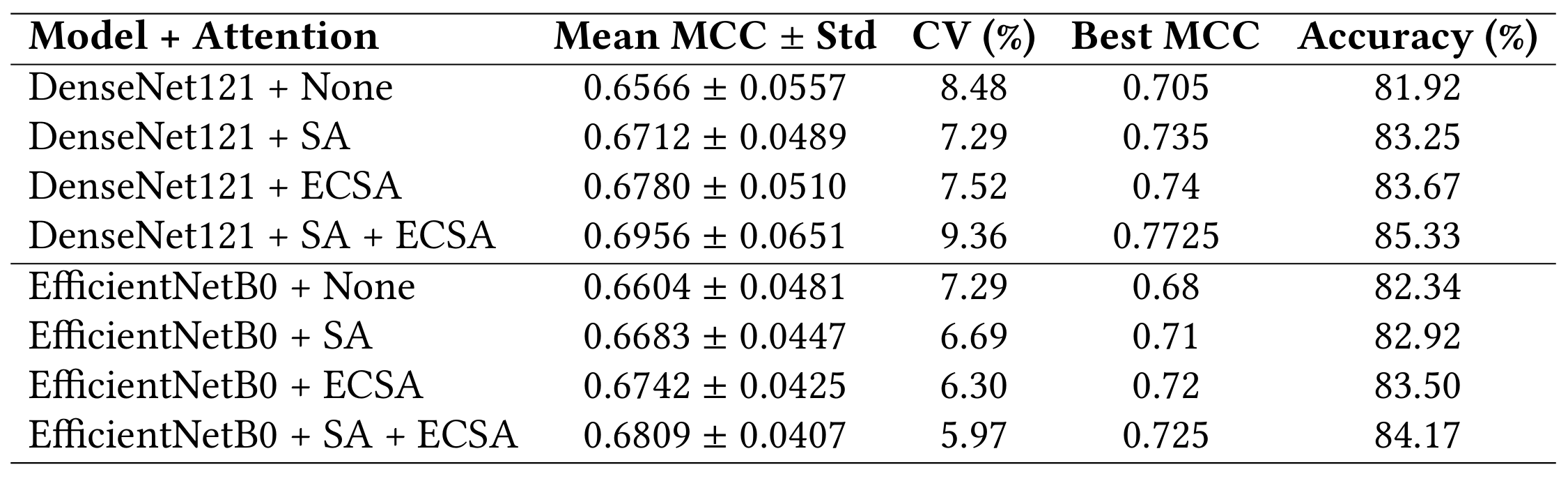
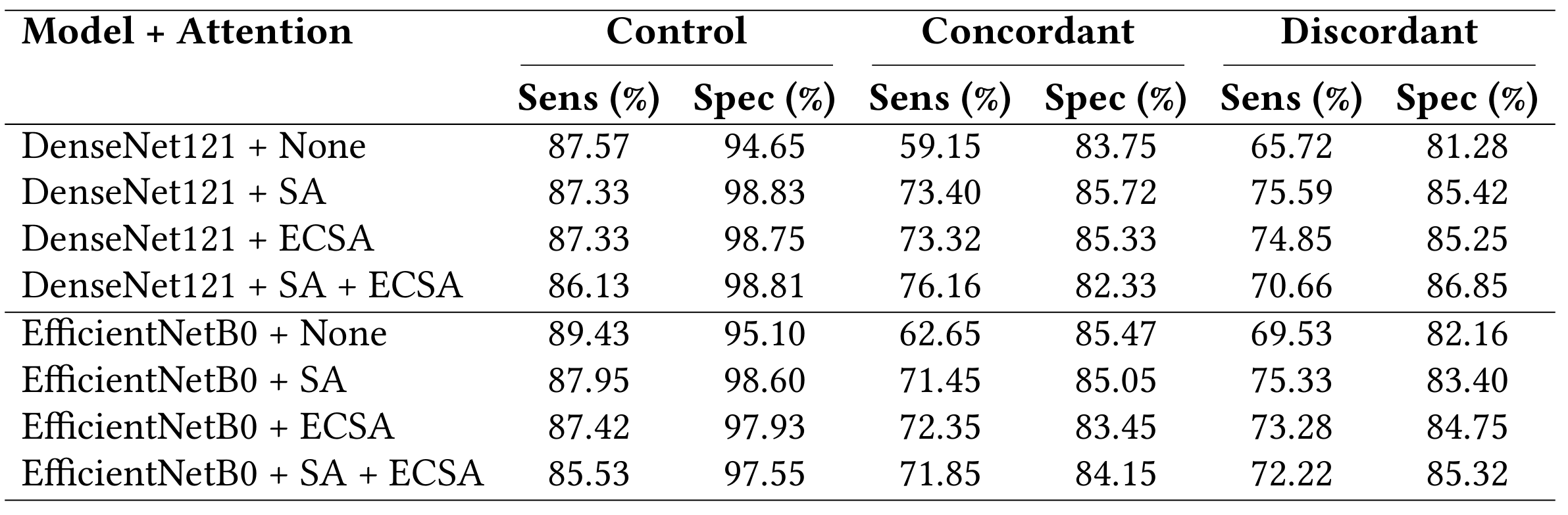
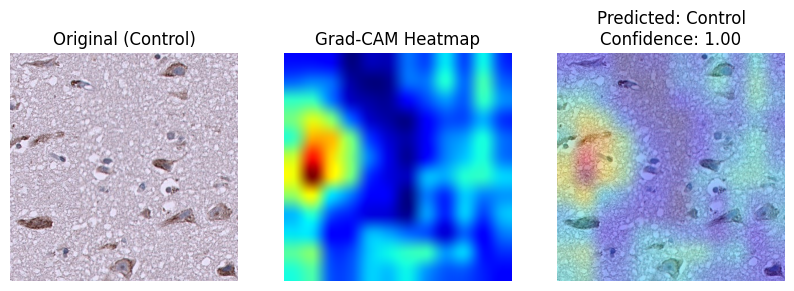
My focus was on evaluating model architectures, attention mechanisms, and explainability techniques to build something trustworthy and informative. I compared DenseNet121 and EfficientNetB0 with combinations of Self-Attention and Effective Channel Spatial Attention (ECSA). The best-performing model (DenseNet121 + SA + ECSA) reached 85.33% accuracy and an MCC of 0.7725. To ensure clinical trust, I used Grad-CAM for explainability and calculated several custom XAI metrics like activation focus and class similarity. The final results show strong performance and reliable interpretability, helping bridge the gap between AI models and clinicians.
Clinicians currently can't distinguish between Concordant and Discordant cases in TDP-43 stained images, which makes this a particularly difficult and important challenge. The goal of this project wasn’t just to classify images, but to help deepen clinical understanding of how ALS presents at the pathological level.
Presenting my research at the Computer Science Poster Day